Background
Acute myeloid leukemia and myelodysplastic syndrome (AML/MDS) represents a broad and complex group of diseases characterized by involvement of the myeloid lineage on hematopoiesis with remarkable variability on clinical presentation, genes mutations, karyotype and molecular abnormalities. Precise characterization on AML/MDS biology is important and the identification of the pathogenic mutated genes is relevant for risk stratification and therapy strategy, mainly for those patients in which targeted therapies is a must. Among pathogenic mutated genes in AML/MDS; TP53 is associated with lower response and high mortality rates. The cumulated number of mutated genes is associated with bad clinical outcomes as well.
Aims
To explore the frequency and impact of mutated genes on clinical outcomes in a cohort of Peruvian patients diagnosed with AML/MDS studied at diagnosis.
Methods
In this report we showed 31 consecutive AML/MDS cases treated at three medical centers in Lima (Clínica Delgado, Clínica Anglo Americana and INEN) from January 2016 until July 2020. Information available at diagnosis was: clinical, karyotype, molecular PCR panel, Illumina TruSight Myeloid Sequencing (TSM) panel which include 53 genes involved on oncogenesis, therapy regimen, response to treatment and overall survival (OS).
Results
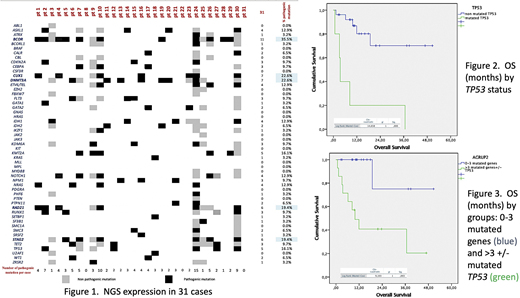
A total of 31 cases were studied. F/M ratio 0.55 (11/20), median age 62 years (range 16-87), number of cases by disease were AML=23 and MDS=8. Among the 26 available karyotypes, the abnormal vs diploid karyotype ratio was 0.18 (4/22). Ad hoc PCR panel were positive in just 5 cases among 30 studies available, for FLT3-ITD(n=2), CBFB-MYH11(n=1), NPM1(n=1) and AML-ETO (n=1). In the TSM analysis, median number of pathogenic mutated genes was 3 (range 0-15). Most common (≥18%) pathogenic mutated genes wereBCOR (n=11; 35,5%), CUX1 (n=7; 22,6%) , DNMT3 (n=7; 22,6%), RAD21(n=6; 19,4%), and STAG2 (n=6; 19,4%)with different rates compare with other series (figure 1). Response rate were as follow: 58,1% complete (18/31), 9,7% partial (3/31), 19,3% non-response (6/31) and 12,9% non-evaluable (4/31).
The estimated median OS was 36,67 months for the entire cohort, the 1 and 3-years OS rates were 69,9% and 62,2% respectively. For AML and MDS cases median OS was not reached, 1-year OS rates were 68,6% and 72,9% respectively, 3-years OS rates were 54,9% and 72,9% respectively (Log Rank 0,071 p=0,79). MutatedTP53was detected in 5 cases (MDS=3 and AML=2) with no therapy response in 4 and all of them had already dead; median OS for mutatedTP53cases was 3,53±0,43 months; 1-year OS rates for non-mutated vs mutatedTP53cases were 80,2% and 20% respectively, 3-year OS rates for cases with non-mutated vs mutatedTP53were 70,2% and 20% respectively (Log Rank 14,6 p<0,0001) (figure 2). The median OS for mutatedBCORwas not reached; 1-year OS rates for non-mutated vs mutatedBCORwere 77,4% and 52,6% respectively; 3-year OS rates for non-mutated vs mutatedBCORwere 64,5% and 52,6% respectively (Log Rank 0,09 p=0,76). The medians OS were not reached for <3 and ≥3 pathogenic mutated genes cases, 1-year OS rates were 92,3% and 53,5% respectively and 3-years OS rates were 73,8% and 53,5% respectively (Log Rank 1,98 p=0,159). The median OS for ≥4 pathogenic mutated genes cases was 11,93±2,5 months, 1-year OS for <4 and ≥4 were 88,9% and 42,4% respectively and 3-years OS for <4 and ≥4 were 71,1% and 42,4% respectively (Log Rank 3,293 p=0,07). Ad-hoc composite analysis for cases of <4 vs ≥4±mutated-TP53showed medians OS not reached and 9,87±2,9 months respectively; 1-year OS rates were 100% and 40,8% and 3-years OS were 75,0% and 40,8% respectively (Log Rank 9,10 p=0,003)(figure 3).
Conclusion
NGS for LMA/MDS is an important tool for diagnosis, assessment and risk stratification in our serie of cases. Only 5 over 53 genes were detected in ≥ 18% of casesBCOR, CUX1, DNMT3, RAD, and STAG. TP53 was related with a high therapy failure response and short OS. Number of pathogenic mutated genes ≥3 or ≥4 by TSM showed tendencies for shorter OS. A composite analysis for patients with 4 mutated genes with or without mutated-TP53showed significant shorter OS. Further studies with mature data and more cases are warrant to propose genetics characterization and risk stratification by NGS in Peruvian AML/MDS cases.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal